Research
Quantitative genomics and computational biology applied to livestock
Our group develops and applies statistical and computational methods to dissect the genetic architecture of economically relevant traits in livestock. We combine large phenotypic datasets with high-throughput genomic technologies such as SNP arrays, whole-genome sequencing, RNA-seq, and DNA methylation profiling to understand the connections between genome and phenotype.

Active Projects
Major funded research initiatives currently underway in our lab
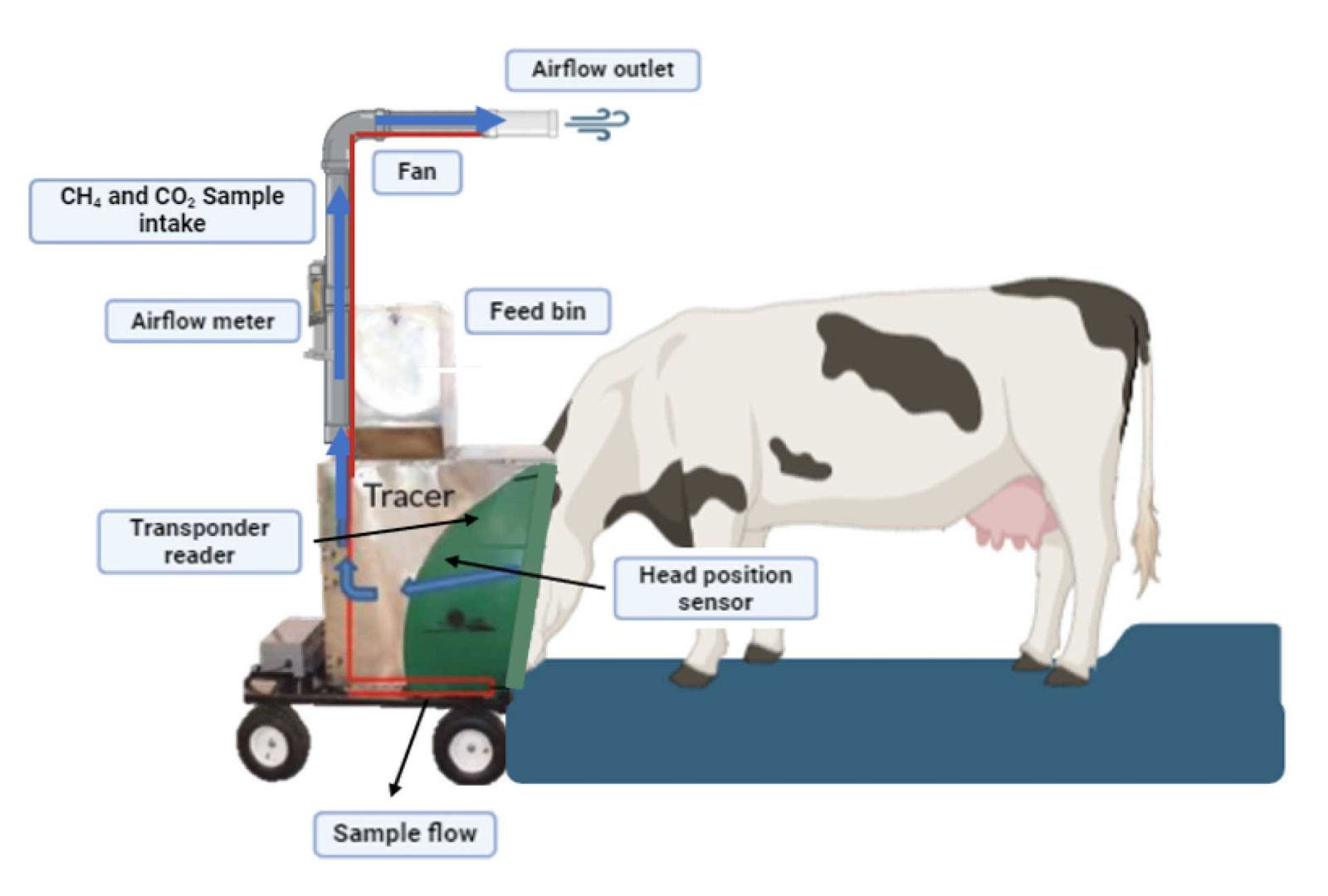
Reducing Enteric Methane Emissions from Dairy Cattle
Integrating genomic, milk spectrometry, and microbial manipulations to mitigate enteric methane emissions from dairy cattle
PI: Francisco Peñagaricano | Collaborators: Michigan State University (MSU), Iowa State University (ISU), University of Florida (UF), University of California, Davis (UC Davis), U.S. Department of Agriculture (USDA), Council on Dairy Cattle Breeding (CDCB)
Why it matters
Methane represents 11% of total U.S. greenhouse gas emissions, and enteric fermentation alone accounts for 27% of total U.S. methane emissions. Enteric CH4 also represents a loss of 6–12% of gross energy intake, meaning that reducing emissions benefits both the environment and animal feed efficiency. This multi-institutional project combines three complementary strategies to tackle the problem.
Selective Breeding
Build a reference population of ~4,000 lactating cows phenotyped at six research farms over three years using GreenFeed systems. Develop genomic evaluations for methane emission traits and incorporate them into national selection indices.
Milk Spectrometry
Use Fourier transform mid-infrared spectroscopy (MIR) to predict CH4 emissions. This high-throughput, low-cost, non-invasive approach can be deployed for large-scale genetic evaluations and on-farm management decisions.
Rumen Microbiome
Characterize the composition and activity of the rumen microbiome in low vs. high methane-emitting cows using metagenomics and metabolomics. Exchange ruminal contents between low and high emitters to assess the relative contributions of host genetics and microbial community.
Lab members involved
Ümit Bilginer, Afees Ajasa, Dérick Rösler, André Zambon, Emily Tabor, Barbara Nascimento, Negin Sheybani
Dairy Cattle Genomics
A major focus of our lab is the genomic analysis of traits affecting animal productivity and welfare. We are particularly interested in fertility, health, feed efficiency, and resilience in dairy cattle. These traits are generally complex, controlled by many genes with small effects, and influenced by environmental factors.
We use genome-wide association studies (GWAS), genomic prediction models, and whole-genome sequencing data to identify genetic variants and biological mechanisms underlying phenotypic variation. Our work on dairy cow resilience, for instance, focuses on developing and evaluating alternative phenotypes that capture an animal's ability to maintain performance under environmental perturbations.
Key questions:
- What genomic regions and biological pathways control fertility and reproductive performance in dairy cattle?
- Can we identify genetic markers for disease resistance and improved animal welfare?
- How can we leverage whole-genome sequence data to improve the accuracy of genomic predictions?
- What are the best phenotypic indicators of resilience, and are they heritable?
Nutritional & Environmental Genomics
Environmental conditions during early development, such as maternal nutrition, heat stress, and other stressors, can have lasting effects on offspring performance through epigenetic mechanisms. Our lab investigates how prenatal and early-life factors alter the epigenome and transcriptome, and whether these changes persist across the animal's life.
We use multi-omics approaches integrating RNA-seq, whole-genome bisulfite sequencing (WGBS), and reduced representation bisulfite sequencing (RRBS) to characterize changes in gene expression and DNA methylation patterns. A central project examines how in utero heat stress affects liver and muscle tissues in dairy calves, linking molecular changes to downstream performance outcomes.
Key questions:
- How does prenatal heat stress reprogram the epigenome and transcriptome of dairy calves?
- Which genes and pathways are epigenetically sensitive to early-life environmental conditions?
- Do epigenetic marks induced by maternal environment persist into adulthood?
- Can we use epigenomic data to predict long-term phenotypic outcomes?
Network Modeling
Complex traits arise from the coordinated action of multiple genes interacting with each other and with the environment. Traditional GWAS approaches treat traits independently, but in reality, traits are often causally interconnected. Our lab develops and applies network-based approaches to understand the relationships between genes and phenotypes.
We use structural equation models (SEM) and causal discovery algorithms to infer gene-phenotype networks in multivariate genetic systems. These methods allow us to move beyond correlation-based associations and identify causal relationships. For instance, we can distinguish whether a genetic variant affects one trait directly or indirectly through its effect on another trait.
Key questions:
- What are the causal relationships among economically relevant traits in dairy cattle?
- Can structural equation models improve our understanding of genetic pleiotropy?
- How can causal discovery algorithms be adapted for high-dimensional genomic data?
- What biological insights emerge from gene co-expression networks in livestock species?
Computational Biology
Modern livestock research generates massive amounts of genomic data that require specialized computational tools for processing, analysis, and interpretation. Our lab develops open-source software and bioinformatics pipelines to address these needs, with a focus on making methods accessible to the broader research community.
Our tools include EnrichKit, which maps genomic coordinates to gene features and performs functional enrichment analysis using biological databases curated for livestock species, and greenfeedr, an R package for processing enteric gas emission data from GreenFeed systems. We also develop analysis pipelines for RNA-seq, WGBS, and multi-omics data integration.
Key questions:
- How can we improve functional annotation of livestock genomes?
- What are the best practices for integrating multi-omics data in livestock research?
- Can we develop standardized pipelines for common analyses in animal genomics?
- How can we make computational tools more accessible and reproducible?
Funding & Support
Our research has been supported by the Foundation for Food and Agriculture Research, the Council on Dairy Cattle Breeding, Gerstner Philanthropies, Dairy Management Inc., and the Wisconsin Dairy Innovation Hub.
